We are a research group in the School of Biotechnology and Biomolecular Science (BABS). We combine bottom-up in vitro synbio from purified proteins, lipids and DNA, with top-down genome editing of microbes. Overall, we apply biophysical methods in synthetic biology and experimental evolution to understand the origins of complex systems.
Research
We study complex systems and their origins, working among microbiology, biophysics, synthetic biology and evolutionary biology. Roughly half of us work, at the moment, on the bacterial flagellar motor. We use this as a model molecular complex to explore protein engineering, directed and experimental evolution, and to understand better how bacterial swimming has evolved and how we can push it to do new things.
Roughly the other half of us work on ‘bottom-up’ synthetic biology where we try to use insights gleaned from understanding an ancient molecular complex (the motor) to build new nanotechnologies out of simple lipid and DNA components to see how we can increase the complexity step-by-step to create new tools in biosensing and light-activation.
We welcome applications to join our group and have spaces for PhD applicants and postdocs where suitable. Please contact Matt if you are interested in joining or collaborating.
Team

Matt Baker

Matt finished an Hons in Chemistry at ANU studying Fluctuation Theorems before completing his DPhil in Physics at the University of Oxford looking at the molecular motor that makes many bacteria swim. There he first learned to love microbiology where you could work with complex living systems but also put them ‘on ice’ when you needed to. Matt started this group at the School of Biotechnology and Biomolecular Science (BABS) in 2018 as a Scientia Fellow. Matt was promoted to Scientia Senior Lecturer in 2019 and to Scientia Associate Professor in 2023. Matt is currently in Athens and Heraklion Greece as a Visiting Professor at the IMBB-FoRTH and the University of Athens. For a more detailed biography see Matt’s UNSW page or go to his linktree to listen to some recent radio.
Pietro Ridone

Pietro joined the lab in July 2019 for his first Postdoc and has since been applying genome editing and synthetic biology techniques to study the stator proteins powering the rotation of bacterial flagella. Born and bred in Florence, Italy, has been fascinated by synthetic biology since the early days of his bachelor years at the University of Glasgow (Scotland, 2008-2012) where he also joined the Glasgow iGEM team for the 2011 edition of the competition. He studied the biophysics of bacterial and human mechanosensitive channels (MscS, MscL and Piezo1) during his PhD in the lab of Prof. Boris Martinac (VCCRI, Sydney), before returning to synbio at the Baker lab. He is currently focusing on engineering the E.coli genome using CRISPR and designing artificial stators to study the evolvability of the bacterial flagellar system. When away from lab duties he can be found roaming around campus in search of strong coffees or defending a goal on the football pitches of Moore park.
Jacob Scadden

Jacob joined the Baker lab in November 2022 after finishing his PhD at the Quadram Institute Bioscience, Norwich, UK (2018-2022) where he studied the regulation of antimicrobial peptides produced by human gut bacteria. Prior to that Jacob studied both his undergraduate degree in biomedical science (2014-2017) and his Master’s in microbiology and infection (2017-2018) at the University of Birmingham, UK. His current Postdoc is focused on ancestral sequence reconstruction of the flagellar filament proteins and its subsequent affect on bacterial motility. Outside the lab, Jacob enjoys hiking and surfing as well as cooking and reading.
Janelle Ramos

Janelle Ramos is a PhD student in the lab who completed Honours in 2021 looking at ancestral reconstructions of the rotor. Janelle commenced a PhD in Feb 2022 researching urinary pathogenic E. coli (UPEC) to examine sequence based diagnostics in catheter based infections.
James Gaston

James Gaston completed Honours in 2021 in the Baker Lab looking at using DNA nanotechnology to colocalise liposomes and quantify this colocalisation. He commenced a PhD in May 2022 in the Baker Lab looking at using DNA nanotechnology to control colocalisation and fusion of liposomes.
Vibhuti Nandel

Vibhuti Nandel started a PhD in March 2022 in the Baker Lab at UNSW. Her research focus is to understand the residues present in the stator complex of the Bacterial Flagellar Motor that control the efficiency of the ion binding. She likes to rejuvenate herself by reading books or following her newfound interest in painting mandalas.
Jamiema Sara Philip
 Jamiema Sara Philip will commence a PhD in our lab in May 2023. She graduated with a Bachelor of Technology in Biotechnology from the University of Kerala in India in 2017. She worked for four years as a research fellow at the Rajiv Gandhi Centre for Biotechnology, India, and then as a project scientist (Bioinformatics) at ICMR-New Delhi, India. Her research studies have primarily focused on understanding bacterial diversity and antibiotic resistance in a variety of environments using metagenomics approaches, as well as SARS-CoV-2 genome analysis. Meanwhile, she completed her Master’s degree in Bioinformatics from Bharathiar University in India in 2022.
Jamiema Sara Philip will commence a PhD in our lab in May 2023. She graduated with a Bachelor of Technology in Biotechnology from the University of Kerala in India in 2017. She worked for four years as a research fellow at the Rajiv Gandhi Centre for Biotechnology, India, and then as a project scientist (Bioinformatics) at ICMR-New Delhi, India. Her research studies have primarily focused on understanding bacterial diversity and antibiotic resistance in a variety of environments using metagenomics approaches, as well as SARS-CoV-2 genome analysis. Meanwhile, she completed her Master’s degree in Bioinformatics from Bharathiar University in India in 2022.
Yara Elahi
 Yara Elahi will commence a PhD in our lab in May 2023. Yara obtained a Bachelor’s degree in Plant Sciences Biology from Kharazmi University, Tehran, Iran. Afterward, he worked on the effectiveness of bacteriophages on clinical multi-drug resistant enterococci during his Master’s thesis. When he graduated with his Master’s in Molecular and cellular Biology-Genetics from Islamic Azad University Tehran North Branch, Iran, he was employed as a research assistant focusing on enterococcal prophages and antibiotic resistance at Tehran University of Medical Sciences. He has also been a research assistant in the R&D Department of Teb Va Salamat Niki Pajouh Company, in Iran. In 2022, he was selected as an editorial board member of Primer Journal which is a subset of the Biotechnology Society of Iran. Apart from the academic side of his life, Yara is a tennis player and a music lover. You can also ask him about the world’s greatest series and movies because watching them is one of his favorite hobbies.
Yara Elahi will commence a PhD in our lab in May 2023. Yara obtained a Bachelor’s degree in Plant Sciences Biology from Kharazmi University, Tehran, Iran. Afterward, he worked on the effectiveness of bacteriophages on clinical multi-drug resistant enterococci during his Master’s thesis. When he graduated with his Master’s in Molecular and cellular Biology-Genetics from Islamic Azad University Tehran North Branch, Iran, he was employed as a research assistant focusing on enterococcal prophages and antibiotic resistance at Tehran University of Medical Sciences. He has also been a research assistant in the R&D Department of Teb Va Salamat Niki Pajouh Company, in Iran. In 2022, he was selected as an editorial board member of Primer Journal which is a subset of the Biotechnology Society of Iran. Apart from the academic side of his life, Yara is a tennis player and a music lover. You can also ask him about the world’s greatest series and movies because watching them is one of his favorite hobbies.
Fatemeh Mohaghegh
 Fatemeh Mohaghegh will commence a PhD in our lab in May 2024. Fatemeh completed her Bachelor of Science in Microbiology from Mazandaran University, Iran. She served as a research assistant in the Biochemistry lab and later engaged as a teaching assistant in Microbiology at Mazandaran University. Thereafter, she worked on the characterization of clonal dissemination and the presence of different virulence factors among methicillin-resistant Staphylococcus aureus strains and obtained her Master’s degree in Medical Microbiology from Isfahan University, Iran. During her Master’s thesis, she collaborated with the Red Crescent Society in Iran, associated with their programs and missions in the field of education and research. Her research studies have mainly focused on bacterial motility, bacterial biofilm and antibiotic resistance.
Fatemeh Mohaghegh will commence a PhD in our lab in May 2024. Fatemeh completed her Bachelor of Science in Microbiology from Mazandaran University, Iran. She served as a research assistant in the Biochemistry lab and later engaged as a teaching assistant in Microbiology at Mazandaran University. Thereafter, she worked on the characterization of clonal dissemination and the presence of different virulence factors among methicillin-resistant Staphylococcus aureus strains and obtained her Master’s degree in Medical Microbiology from Isfahan University, Iran. During her Master’s thesis, she collaborated with the Red Crescent Society in Iran, associated with their programs and missions in the field of education and research. Her research studies have mainly focused on bacterial motility, bacterial biofilm and antibiotic resistance.
Sahiba Shah
Sahiba is undertaking Hons in 2024 in our group.
Ed Luo
Ed is undertaking Hons in 2024 in our group.
Jessica Do
Jessica is undertaking an Engineering Undergraduate Thesis in 2024 in our group.
Oliver Taig
Oliver is undertaking Hons in 2024/2025 in our group.
Alumni
Gonzalo Peralta (Hons 2016)
Dr Yu-Wen Lai (Postdoc 2017, now University of Sydney)
Jessica Clark (Hons 2018)
Oskar Jaggers (Hons 2018)
Es Darley (Hons 2019, now University of Tokyo)
Joon Bae (Hons 2020)
Angela Lin (Biomedical Engineering 2020, now Children’s Cancer Research Institute)
Md Sirajul Islam (Postdoc 2022, now in industry with Cleanaway Waste Management
Imogen Kelly (Hons 2022)
Sehhaj Grewal (Computer Science & Engineering 2022, now Google)
Imtiazul Islam (PhD 2023, now postdoc Nationwide Children’s Hospital, Ohio, with Gabriel Cara-Fuentes)
Divyangi Pandit (Hons 2023)
Alex Mason (Postdoc 2021-2023, now PI at University of Wollongong
Jyoti Gurung (PhD 2024, now postdoc Imperial College with Yuval Elani
Madhuri Sande (Postdoc 2023-2024, now at Macquarie University)
Shruthi Annamalai (Hons 2023-2024)
Projects
We have the following research projects currently active in the lab and open for motivated prospective PhD students to join. Those interested in applying should consider the key dates and application process.
Project Area 1: Ancestral Reconstruction of the Bacterial Flagellar Motor
 We recreate microbial ‘Jurassic Parks’ by resurrecting ancient flagellar motor componentry in contemporary hosts and measuring how well they work to let bacteria swim. This allows us to create ancient motors that have never existed in the present day to synthesise and evolve new motors as well as to learn about the process of evolution. We focus in particular on the proteins that make up the stator complex (the engine) and the filament (the propellor).
We recreate microbial ‘Jurassic Parks’ by resurrecting ancient flagellar motor componentry in contemporary hosts and measuring how well they work to let bacteria swim. This allows us to create ancient motors that have never existed in the present day to synthesise and evolve new motors as well as to learn about the process of evolution. We focus in particular on the proteins that make up the stator complex (the engine) and the filament (the propellor).
Project Area 2: The Origins of Motility.
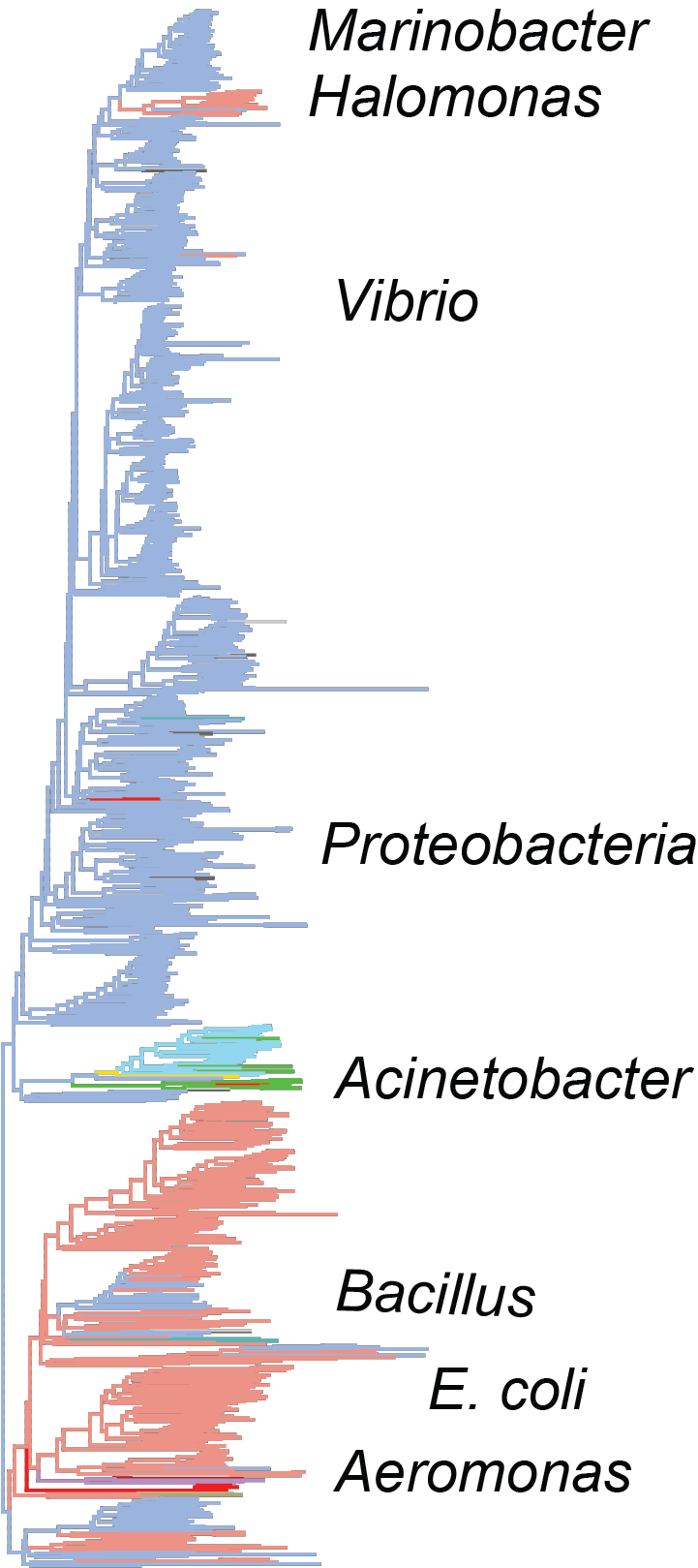 The evolutionary origins of the bacterial flagellum have been a subject of scientific and public controversy – how can evolution produce such a complex system? We make progress on the issue by updating old phylogenetic work with new datasets and improved models and combining this with experimental evolution work being done in our lab. The project is to assemble a well-organized database of flagellar proteins and explore sequenced bacterial genomes with genome browsers and sequence-similarity searches.
The evolutionary origins of the bacterial flagellum have been a subject of scientific and public controversy – how can evolution produce such a complex system? We make progress on the issue by updating old phylogenetic work with new datasets and improved models and combining this with experimental evolution work being done in our lab. The project is to assemble a well-organized database of flagellar proteins and explore sequenced bacterial genomes with genome browsers and sequence-similarity searches.
Project Area 3: Evolution Across Interfaces
We aim to explore how motility evolves across interfaces, when a bacterium faces a change in environment and how bacteria adapt to dwindling nutrient across such an interface. This project has scope for designing and building custom tanks to optimise bacterial evolution using 3D printing and prototyping, as well as investigating microbiology and bacterial motility in multiple dimensions using layered swim devices.
Project Area 4: Driving Fluidic Flows with the Bacterial Flagellar Motor
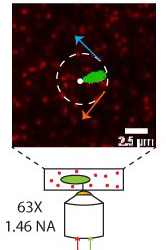 We utilise the high efficiency and self assembly of the flagellar motor to drive rotation of cells on patterned surfaces to control mixing and fluid flows in microfluidics. This project suits someone with an interest in DIY/maker culture and the fabrication of fluidic devices to manipulate and respond to flows. We engineer bacteria to drive rotation only when illuminated by different wavelengths of light and then incorporate external light-based control of flagellar motors to drive microfluidic flows.
We utilise the high efficiency and self assembly of the flagellar motor to drive rotation of cells on patterned surfaces to control mixing and fluid flows in microfluidics. This project suits someone with an interest in DIY/maker culture and the fabrication of fluidic devices to manipulate and respond to flows. We engineer bacteria to drive rotation only when illuminated by different wavelengths of light and then incorporate external light-based control of flagellar motors to drive microfluidic flows.
Project Area 5: Developing DNA-lipid nanotechnology
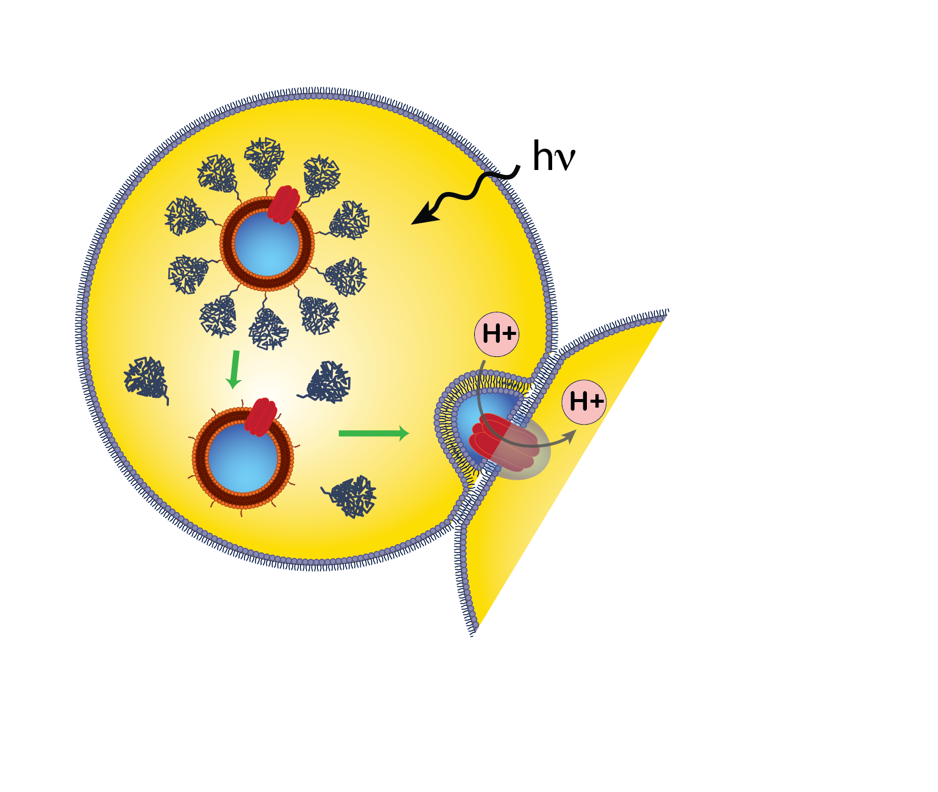 We design DNA origami nanostructures that can bind and release from liposomes to direct the fusion of liposomes. We hope to apply this to trigger liposome fusion on demand using light and electrical signals. This project involves in vitro synthetic biology, DNA and lipid nanotechnologies and microscopy, and has potential for students interested in learning how to build custom high resolution microscopes.
We design DNA origami nanostructures that can bind and release from liposomes to direct the fusion of liposomes. We hope to apply this to trigger liposome fusion on demand using light and electrical signals. This project involves in vitro synthetic biology, DNA and lipid nanotechnologies and microscopy, and has potential for students interested in learning how to build custom high resolution microscopes.
Project Area 6: Signalling across membranes
We use droplet interface bilayers and droplet hydrogel bilayers to create custom membranes of any composition and then a combination of in vitro protein and DNA nanotechnology to add complexity to these systems one component at a time. We aim to make the biological equivalent of the transistor - leveraging logic gates and computational power of DNA nanotechnology with varied protein functions in our synthetic droplet systems.
Project Area 7: Machine learning applications to melanoma diagnosis
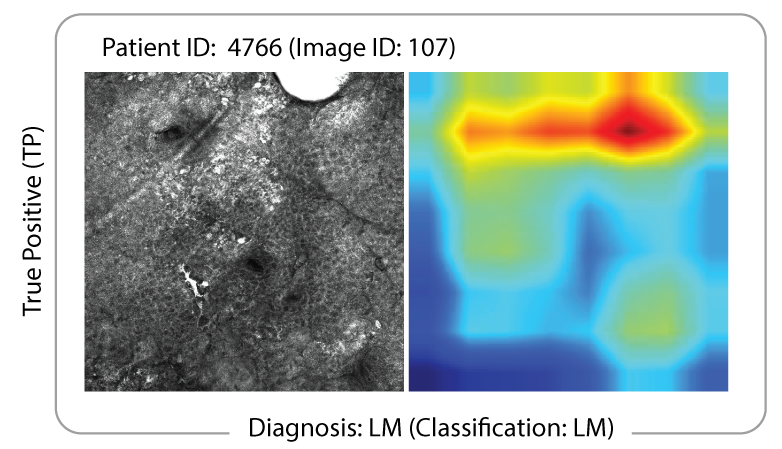 We use machine learning frameworks to classify melanoma images from in vivo and ex vivo confocal microscopy in partnership with our clinician collaborators and A/Prof Fatemeh Vafaee. In our lab we use our knowledge of microscopy and image analysis to optimise data flows for machine learning to then diagnose cancers and detect melanoma boundaries in tissue.
We use machine learning frameworks to classify melanoma images from in vivo and ex vivo confocal microscopy in partnership with our clinician collaborators and A/Prof Fatemeh Vafaee. In our lab we use our knowledge of microscopy and image analysis to optimise data flows for machine learning to then diagnose cancers and detect melanoma boundaries in tissue.
Recent Papers
2024
Hybrid Exb/Mot stators require substitutions distant from the chimeric pore to power flagellar rotation
Journal of Bacteriology
Ridone, Baker.
| bioRxiv |
DIB-BOT: An open-source hardware approach for high throughput droplet interface bilayer deposition
Advanced Materials Interfaces
Mason, Wickham, Baker.
| bioRxiv |
The parasitic lifestyle of an archaeal symbiont
Nature Communications
Hamm, Liao, von Kügelgen, Dombrowski, Landers, Brownlee, Johansson, Whan, Baker, Baum, Bharat, Duggin, Spang, Cavicchioli.
| journal |
Light control in microbial systems
International Journal of Molecular Sciences
Elahi, Baker.
| journal |
2023
Tuning the stator subunit of the flagellar motor with coiled-coil engineering.
Protein Science
Ridone, Winter, Baker.
| journal |
Ion-Powered Rotary Motors: Where Did They Come from and Where They Are Going?
International Journal of Molecular Sciences
Nandel, Scadden, Baker.
| journal |
Microbial stir bars: Light-activated rotation of tethered bacterial cells to enhance mixing in stagnant fluids.
Biomicrofluidics
Gurung, Navvab Kashani, de Silva, Baker.
| journal |
Ancestral reconstruction of the MotA stator subunit reveals that conserved residues far from the pore are required to drive flagellar motility.
microLife
Islam, Ridone, Lin, Michie, Matzke, Hochberg, Baker.
| journal |
Computer-aided diagnosis of reflectance confocal images to differentiate between lentigo maligna (LM) and atypical intraepidermal melanocytic proliferation (AIMP).
Mandal§, Priyam§, Chan, Gouveia, Guitera, Song, Baker•, Vafaee•.
Cancers
| journal |
2022
Building programmable multicompartment artificial cells incorporating remotely activated protein channels using microfluidics and acoustic levitation.
Li, Jamieson, Dimitriou, Xu, Rohde, Martinac, Baker, Drinkwater, Castell, Barrow.
Nature Communications
| PDF | journal |
The rapid evolution of flagellar ion selectivity in experimental populations of E. coli.
Ridone, Ishida, Lin, Humphreys, Giannoulatou, Sowa, Baker.
Science Advances
| PDF
| journal |
2021
Binding of DNA origami to lipids: maximizing yield and switching via strand displacement.
Daljit Singh§, Darley§, Ridone, Gaston, Abbas, Wickham•, Baker•.
Nucleic Acids Research
| journal |
Fluorescence Approaches for Characterizing Ion Channels in Synthetic Bilayers.
Islam, Gaston, Baker.
Membranes
| journal |
Novel Amiloride Derivatives That Inhibit Bacterial Motility across Multiple Strains and Stator Types.
Islam•, Bae•, Ishida•, Ridone, Lin, Kelso, Sowa, Buckley, Baker.
Journal of Bacteriology
| journal |
Pushing the super-resolution limit: recent improvements in microscopy below the diffraction limit.
Nieves, Baker.
Biochemical Society Transactions
| journal |
Flagellar export apparatus and ATP synthetase: Homology evidenced by synteny predating the Last Universal Common Ancestor.
Matzke, Lin, Stone, Baker.
BioEssays
| PDF
| journal |
Earlier and other collaborative publications can be viewed on Matt’s Google Scholar.
Contact
School of Biotechnology and Biomolecular Science
Find us:
Biosciences
Room 301 (Level 3) D26
UNSW Kensington Campus
Send something:
Attention: Matt Baker
BABS
Upper Campus Store E26, Bioscience South
LG018 Loading Dock
Via Gate 11, Botany Street
UNSW Sydney NSW 2052
Australia